It's the hottest new biotechnology technique to hit the headlines since. well, since ever. CRISPR is a precision set of genome editing tools enabling scientists to cut and paste together DNA in any organism, exactly how they want - and the implications for human health, and even humanity, are huge. Plus, linking genetics to lifestyle, and our gene of the month is black and white. and very cute.
In this episode
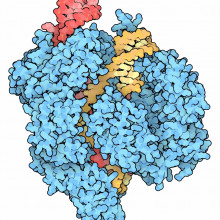
01:10 - Nell Barrie - History of CRISPR
Nell Barrie - History of CRISPR
with Nell Barrie, Science writer
Kat - The speed at which CRISPR has moved from the pages of dry academic journals to the headlines of newspapers - not to mention labs and biotech companies around the world - has been breathtaking. This month the UK Human Fertilisation and Embryology Authority granted a licence for scientists at the Francis Crick Institute in London to start using the technique to modify early-stage human embryos - although these would not be allowed to grow past the stage of being a tiny ball of cells.
Although it might seem like the CRISPR story has come out of nowhere in the past year, the Naked Genetics podcast was on the case back in 2012, when University of California, Berkeley researcher Jennifer Doudna and her collaborator Emmanuelle Charpentier, who was at Umea university in Sweden, published a paper in the journal Science entitled "A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity". Here's a clip from the podcast back in July 2012, where science writer Nell Barrie and I discuss the implications of their discovery.
Kat - I love this just because of the potential of the story, and this is researchers who've been looking at bacteria and looking at how bacteria use certain molecules like molecular scissors to snip up their DNA and glue it together, and it helps them develop different characteristics to help them survive. But they've actually looked at these molecular scissors and found out how they work. And they think that possibly, you could use this technology to make programmable scissors so you could cut up human DNA, animal DNA, and basically glue it back together in any kind of way you like.
Nell - That's pretty cool. I mean, I think it's kind of what I think of as genetic engineering. I guess you think you'll just put together whichever bit of the DNA that you want, but actually, in reality, that's not really possible yet. So I suppose this is taking us a little bit closer to real designer genetic tailoring or whatever you might like to call it.
Kat - So I remember when I was in the lab - and this is some time ago so things may have changed - but if you wanted to do genetic engineering and stick different bits of DNA together, you had to look at the sequence and there were only certain sequences that you could cut using enzymes. Things have probably moved on now, but I think there is some restriction in the sequences that you can cut and glue together. So having a different set of technology could really be exciting.
Nell - Yeah, and I guess that sort of eventual implications would be that you could be create these kind of designer organisms like maybe, bacteria that degrades nasty environmental toxins, or produce things that we need for drugs, or all kinds of exciting applications. Because we know that bacteria and fungi have all these abilities and it's just about, could we harness them to do the things we want, I guess.
Kat - Watch this space, I think because it is still very early days for that, but thank you very much. That's Nell Barrie, Science Writer.
03:53 - Tony Perry - What is CRISPR?
Tony Perry - What is CRISPR?
with Tony Perry, University of Bath
Kat - Let's take a closer look at exactly what's involved in CRISPR - or to give it its full name, CRISPR/Cas9. To find out more about this molecular toolkit and what it can do, I went along to a meeting organised by the Progress Educational Trust, discussing new genetic techniques and the impact they could have on human health and human genomes in the future. I caught up with one of the speakers, Tony Perry from the University of Bath, who told me more about the story of CRISPR, and some of its potential applications.
Tony - The prototype comes from bacteria that give you a sore throat, actually. It's sore throat bacteria Streptococcus pyogenes. It seems to be a kind of defence system and it turns out that even bugs get bugs. And so, these Streptococcus pyogenes, these sore throat bacteria themselves get invaded by viruses. The CRISPR/Cas9 was evolved as a way of cutting the viral DNA and thereby defending themselves, the Strep pyogenes, against these viral invaders.
Kat - So, what are the components of this self-defence system? How does it work and how have researchers then hacked this to do more precise editing?
Tony - The system really is like a pair of molecular scissors and a molecular satnav. The satnav is a small RNA molecule and it guides the molecular scissors to precisely where you want it to go in the genome. The reason it does that is because like a satnav, it's programmable. So you get to tell satnav what the coordinates are in the genome and it will then take the scissors to that point in the genome and the scissors will cut there. When they do so, when the scissors do so, they make a double-stranded break called the DSB and then the cell has got two types of tool kit that it can use to repair that double-stranded break. One type of repair that the cell can use or one tool kit is a simple kind of paste mechanism...
Kat - Just get the ends and glue them back together.
Tony - Absolutely, just get the ends and glue them back together again quickly. It does have to be a rapid process because these breaks are anathema to cells. If they're not repaired quickly, the cell can die and even worse, it can give rise to cancer and death of the organism in multicellular organisms. Well, that's very useful for research because you can use it to knockout genes if the cut is in a gene. It's probably not going to be so useful for clinical applications anyway. The second toolkit that the cell has or mammalian cell has to repair a double-stranded break is called homology-directed repair. We can actually add in our own designer piece of DNA which is used for repair. And so, we can design that, produce it, fabricate it in vitro in the laboratory and make sure by sequencing that is exactly what we want. And then we can introduce it into the cell of the same time as we've introduced the molecular satnav and the molecular scissors, the CRISPR/Cas9. And so, the cell in the right circumstances will use this extra piece of DNA that we've added as a repair template. What you end up with is in effect this sequence that you've added precisely inserted in the targeted position of the genome so that it's been used to repair the genome and you've introduced change that corresponds to the DNA that you've added in.
Kat - So, we've got the CRISPR which is the RNA, the guiding system, we've got the Cas9, the scissors. And then this DNA template, and that could be anything I can imagine.
Tony - Can't quite be anything you can imagine as yet. Perhaps one day. One constraint at the moment is the size if you want to introduce a piece of DNA. So, you make the cut where you want the cut to be. So, you can introduce a large piece of DNA where you want it to be. But there probably is an upper limit at the moment today on how much novel DNA you could introduce. We're probably in the many thousands of base pairs or many thousands of characters in the genome and we don't know whether the sky is the limit or not. But what we do know is that as you increase the length of information that you want to introduce, the efficiency drops. And so probably, if you wanted to introduce a really large piece of DNA for whatever reason, you might have to do that in multiple steps.
Kat - So, we've got this technology which seems like an incredibly powerful tool and I know that scientists all over the world are using it in all kinds of ways in research in the lab, to understand how genes work and how genes get turned on and off, and all this kind of thing. But what we're talking about at this meeting here is editing the human genome. Tell me about the two different approaches that people are trying to think about - not doing necessarily at the moment - but the two sort of different ways that people are thinking about doing this at the moment.
Tony - As I see it, this whole topic can be sort perhaps rather artificially but sorted into whether the changes that you make to the genome are heritable - in other words, they can be passed on to successive generations - or are not heritable. So, the two main types of non-heritable genome editing changes that you might make work together with existing technologies that are variously well-established.
Kat - So this is effectively just normal cells of the body that are broken. You're repairing them in some way.
Tony - You can repair cells in situ using gene therapy and this would be coupled together very nicely with CRISPR/Cas9 that you here are doing the editing as it were of the patient's cells in situ. And there are one or two clinical trials that are already in progress to use this kind of in situ approach of somatic cell gene therapy.
Kat - What sort of diseases are people starting to look at?
Tony - Well, the ones that I am aware of are for example retinopathies. So, you can go in and you can edit genes that predispose to certain types of retinopathy.
Kat - That's problems with eyesight - the back of the eye.
Tony - Exactly, problems with eyesight and this is quite a good place to start because the eye is relatively accessible and a great deal is known about the eye. And so, this is something which lends itself to this application of CRISPR/Cas9. So, this is an example, perhaps the first of many, but it's an example of somatic cell gene therapy combined with CRISPR/Cas9. So, somatic cell gene therapy at the moment, there are probably over 600 clinical trials on-going in the US and it's a technology that's been around for some 25 years or more since the first successful somatic cell gene therapy. So, we might see this with the CRISPR/Cas9 which is very new, being combined with very well-established clinical technologies.
The second non-heritable type of application is a cell therapy where you take cells from a patient who's affected with a particular genetic condition that predisposes to a disease. And you then edit those cells in vitro in the laboratory and then you can confirm that you've made the edit that you want to make. And you can put those cells in effect, you can put them then back into the patient because you've now fixed as it were the genetic change that predisposed to their condition. And that can happen either by taking cells and directly editing them and putting them back in - this is largely speculative although there are one or two on-going pieces of work. The applications for that are for example, where you can recover hematopoietic cells from the bone marrow of patients. And then you can potentially edit genes in these blood stem cells and then transplant this bone marrow back into the patient so that now the patient's bone marrow cells have got their repaired version of the gene, and the problem is fixed.
Another way, perhaps a little bit more futuristic but is conceivable, is combining CRISPR/Cas9 with the whole technology of induced pluripotent stem cells. So induced pluripotent stem cells was something that was discovered by a method that was reported by Shinya Yamanaka. He won the Nobel Prize in 2012 for this. Basically, what you can do is you can take somatic cells like skin cells and if you do the right treatment, if you subject them to the right factors, you can cause them to start behaving like embryo-like cells. From those embryo-like cells, you can ask them to then respecialise into whatever cell you ask them to specialise into. So now, we have a system if we combine this iPS cell technology where we could, if we have a patient - perhaps a patient with a neurological disease that has a genetic predisposition - we could take skin cells from that patient because although skin is not the brain clearly, those cells in many cases would also carry the same mutation. It's just that there's no manifestation of the mutation there because they're not neurons. But we can take skin cells, generate from them iPS cells, embryo-like cells, we can go in with CRISPR/Cas9 - so combining the CRISPR/Cas9 here - fix the problem and then ask these embryo-like cells - these iPS cells - to differentiate to become neurons which we can do in many cases in many different cell types. And then we can transplant the neurons which are derived in effect - it's a bit around about but they are derived from the patient - back into the patient. But now, those neurons have got the fixed genome. So, that's another example of non-heritable because the neuron is not the germ line. So, that's another example of how CRISPR/Cas9 might be used in non-heritable therapy. Not CRISPR/Cas9 but this kind of iPS approach is being used, has been used in clinical trials for example in Japan.
Kat - Tony Perry from the University of Bath. And we'll be hearing more from him in next month's Naked Genetics podcast, when we take a look at whether CRISPR could be used to genetically engineer humans, and - getting to the heart of the burning questions - I ask whether my friend could hack her genome to grow a tail.

14:58 - Azim Surani - Should we edit humans?
Azim Surani - Should we edit humans?
with Azim Surani - University of Cambridge
Kat - Back in December, the US national academies of sciences and medicine, the Royal Society in London and the Chinese Academy of Sciences convened a major summit in Washington to discuss the use and implications of using CRISPR for modifying human cells and embryos. The technology to do this is either in place or in progress, but just because we can, doesn't mean that we should.
Professor Azim Surani from the Gurdon Institute in Cambridge went along and I spoke to him during a break at the Progress Educational Trust conference - where he'd been presenting his work on creating egg and sperm cells from somatic cells in the body such as skin - to find out who went to the Washington summit, and what happened.
Azim - Well, there were scientists and there were ethicists, there were lawyers. So, it was a mixed group of people. So the scientists were putting the information on and how far the technique has got and what new developments might come.
Kat - Presumably, what we don't know as well and what some of risks are.
Azim - There are risks. For example, most of the work suggests that they don't have particular problems with off target effects.
Kat - This is accidentally cutting things you don't want to cut?
Azim - Yeah. It looks like the incidence of off-target effects is far less than was anticipated. So, that's one good thing.
Kat - In terms of the technology, we now know that there are researchers who are working on how to take cells from the body - somatic cells like skin cells and then treat them in certain ways, and even potentially turn these into egg and sperm. I mean, that's a whole world - not needing to take an egg and a sperm from a man and a woman and edit them. But actually, you could just take someone's skin cells and make a baby and change those genes.
Azim - Because these are children that are not born yet so you can't talk about informed consent.
Kat - So kind of the playing God argument.
Azim - And then other people say, yeah, well when you have children naturally you don't ask their permission. You're just passing on your genes.
Kat - I didn't want to be born!
Azim - Good and bad. You don't go and say, "I didn't get permission from the children I gave birth to" or something like that. So, those are things that did come up at the meeting. I think the thing that was surprising to me was that I went there thinking that there'd be a lot of people who would be saying, "Gene editing, germ line gene editing is a red line. We don't cross this. I found that overall, people were much more prepared to debate this and they didn't say, "We should never do it." The consensus at the end was that we should review this periodically so they want to see how the science is progressing, what the public views are, whether the public opinion is shifting like it did for some of the other things.
So, I think that's a good thing because I think we have to see - so, I think at the moment, the decision is that people just press on. They're doing basic research. So, if you have for example a very good reason why you want to use CRISPR/Cas9 on human embryos for example, not to implant but, to understand how genes work, how cells are made, and so on. So, I think the general consensus is that this should be allowed. I think this could also be the case with cells that give rise to germ cells. I've already used it in the synthetic germ cells we are making to understand how genes are working.
Kat - How important is it, this isn't just a decision that the scientists make? Say, "Okay, we understand the science. We should be the ones to decide how this works."
Azim - I think that absolutely not. I don't think that scientists are forcing this issue. I think my reading of the situation is that scientists are excited about the work they're doing because I think this is a very powerful tool and so, most of the basic researchers see this as an opportunity to gain knowledge. That's exciting enough for them. They're not necessarily interested in pushing this onto the public and say, "Now, we need to decide because we can do it. we will do it." I think the overall, the tone of this meeting was that we will present the signs and we want to inform the public and then it has to be a decision for the public through the regulatory committees and agencies. They differ in different countries so it's not going to be uniform. That was very clear that it's not - it may be that the regulatory framework is not going to be the same everywhere because there are different conditions and things like that. So, it will very much be dependent on each country, kind of taking this on, but doing it through this interaction, international interaction so that there's a debate internationally as well.
Kat - And to return to some of the more exciting technology down the road, people are now starting to talk about making egg and sperm cells from cells in the body, potentially engineering those. How do you see that kind of technology unfolding? Is this going to be the future of this kind of technology?
Azim - I think we are - to be honest - we are far from that. I think really, we're just taking the first steps. I mean, we're just literally being able to make the very early germ cells. Also, a much more complicated issue is whether they're going through proper programming. You'd have to see whether they're going through all of those kinds of changes properly in culture.
Kat - That was Professor Azim Surani from the Gurdon Institute in Cambridge.

21:28 - Suzi Gage - Linking genes to lifestyle
Suzi Gage - Linking genes to lifestyle
with Suzi Gage, University of Bristol
Kat - With all this talk of engineering the genome, it's important to remember that we really don't understand much at all about how our genes work and what they do. Every year scientists find hundreds if not thousands of associations between tiny variations in our DNA and the risk of all kinds of diseases and disorders from cancer and heart disease to diabetes and depression through Genome Wide Association Studies, usually known as GWAS. Unpicking exactly how these genetic variants work is a complex task, and a new paper from Suzi Gage and her colleagues at Bristol University has just thrown some more complexity into the mix - the impact of genes on lifestyle factors. I asked her to explain what she's found.
Suzi - The sort of rationale behind this paper is just to get a better handle on what the results of genome wide association studies actually mean, what they can actually tell us. Because it's all well and good knowing that certain genetic variants are associated with a disease or with a phenotype of any kind. But until you actually can sort of delve a bit deeper and work out what that association actually means that's only of have limited interest really.
Kat - So, we hear these stories in the papers. Ten new genes for cancer found, hundred new genes for autism. Is that this kind of study that's throwing up these genes and we don't really know what they are and what they do?
Suzi - Yeah, absolutely. So, in some cases, you do know what a particular genetic variant does in terms of coding for a protein or something like that where you can actually test it. But more often than not actually, we don't know at the moment and so, the question is, how do we know what these variants are actually doing? Is it some sort of biological effect that they're having on the disorder - on cancer for example - or is it actually that they're having an effect on something else and that something else might be having the effect on cancer?
Kat - So, unpack that a bit. What do you mean by that? What examples would apply here?
Suzi - The reason that we became interested in this is that we noticed that this genetic variant that is quite robustly associated with how heavy a smoker you are, if you are a smoker, this genetic variant was actually seen in a genome wide association study of lung cancer. So, that could mean that there's just some sort of shared genetic architecture - so this gene might have an effect on both smoking and your risk of lung cancer. But what seems a far more parsimonious explanation of this is actually that the reason that this genotype or this genetic variant is coming out of the lung cancer GWAS is because smoking causes lung cancer. So genetic variants that predict smoking are going to be seen in GWASs of lung cancer.
Kat - Basically, if someone has this genetic variation and they are a smoker, they're going to want to smoke loads and that's what's going to be more likely to give them lung cancer.
Suzi - Exactly and that increases their risk of lung cancer. Absolutely, yes.
Kat - So, are there any other genetic variations that might be working in a similar way?
Suzi - Quite possibly, yes. At the moment, the sort of main thing holding us back is actually, we don't know the specific genotypes that are associated with these kind of modifiable exposures. There's a good example in the alcohol literature. There's a genetic variant that quite strongly predicts alcohol consumption. It's not very common in European populations but a bit more common in samples of east Asian ancestry. If you look in these populations then genome wide association studies of high blood pressure identified this particular alcohol-related genotype. But it's not seen in GWASs of high blood pressure in European populations which is pretty strong or certainly fairly strong evidence that it might be alcohol causing high blood pressure. And that's why you only see this particular genotype in these GWASs in these specific populations.
Kat - Do you think that there might be other ones out there? We've done cigarettes and alcohol. Any other vices that might be linked to conditions?
Suzi - Well, absolutely. Quite a lot of substance use is known to be heritable- so for example, cannabis use. But as yet, we haven't identified the specific genetic variants that are associated with cannabis use. But as and when these variants were identified, it would be perfectly plausible that we might see these genetic variants coming out of GWASs for diseases. So, if there is a causal link between cannabis and schizophrenia. We identified this cannabis risk-increasing genetic variants. Then if there's truly a causal association between cannabis and schizophrenia, you might well expect that these genetic variants would come out in GWASs of schizophrenia.
Kat - So, what do we do with this information? I always find it interesting with these genetic studies whether you sort of say, "Oh well, you've got the gene variation that makes you more likely to be a stoner. Well, what do we do?" Or you're going to smoke more so, bad luck? Where do we take this work?
Suzi - In terms of what these studies might actually tell us, when we're looking at genome wide association of diseases, if a genetic variant comes out of one of these studies, that's actually telling us that there's a modifiable risk factor that's increasing the risk of this disease then that's a much easier place to target and intervention at than a biological mechanism.
Kat - So, it's easier to help someone stop smoking say, than it is to find and develop a drug that targets a molecular pathway that might reduce their lung cancer risk.
Suzi - Absolutely, yeah.
Kat - But don't we already know that people should give up smoking?
Suzi - Well, I mean, that is a very good point. I think being able to sort of say a bit more definitively than what we're seeing are causal associations here can only help in terms of getting that message across.
Kat - There's also a lot of talk about the nature versus nurture or nature with nurture and I guess what you are unpicking here is how our genes interact with the things that we do, the things in our lifestyle. It feels like this is really first steps into that world - that we still don't really understand much about how our lifestyle and environment affects our genes and what happens to us?
Suzi - Yeah. I think that's the key point here that we have to be very cautious when we kind of interpret these studies because we're really just at the very beginning of being able to understand what these genetic associations might actually mean and how we can then harness them to improve people's health and improve our own understanding of humans, our behaviour, and our health.
Kat - Bristol University's Suzi Gage, and her paper was published this week in the journal PLoS Genetics.

28:26 - Gene of the Month - Panda
Gene of the Month - Panda
with Kat Arney
And finally it's time for our Gene of the Month, and this time it's the cute and cuddly Panda. Rather than being a gene in these famous black and white bears, Panda is found in sea urchins and is short for paracentrotus anti-nodal dorsal activity. It encodes a molecule that's important for helping a developing sea urchin embryo tell its front side from its back - known as dorso-ventral patterning - and is laid down in the sea urchin egg before it's even fertilised. Panda works by acting against another important developmental molecule called nodal, which tells cells in the developing urchin to become ventral (or the front bit) rather than dorsal (that's the back).










Comments
Add a comment