Around one in four adults in the UK experiences mental health problems in any year, and mental illness is a major problem for sufferers, their loved ones, and society. It's becoming clear that genes are involved, but what do we actually know? Plus, artificial chromosomes, autism in the womb, and a toddling little gene of the month.
In this episode

01:03 - Prof Cathryn Lewis - Depression genes
Prof Cathryn Lewis - Depression genes
with Professor Cathryn Lewis, Kings College London
This month I'm reporting back from the Genetics Society Spring meeting, which was held at the beginning of April at the Royal Society in London. Called "Psychiatric Genetics: Pathways and Prospects", the meeting featured talks from leading scientists from around the world, discussing how genetic research is shedding light on mental illness and improving life for sufferers.
To kick off, I spoke to Cathryn Lewis, professor of statistical genetics and epidemiology at Kings College London, who's searching for genes involved in depression. I asked her to explain a bit about the scale of the problem when it comes to psychiatric diseases in the UK.
Cathryn - Mental health issues in general, and depression in particular, are a major issue in the UK. So, up to one in five people will suffer from depression at some point in their life and for some people, that will be a major impact on their life. They will have recurrent episodes of depression throughout their life, and never really recover their full life as it was before they became depressed. And of course, that has an enormous impact on individuals' lives, on their family, it causes problems for employment and security of life, and economically, it has a major effect on the UK economy.
Kat - When we talk about depression, we're not talking about feeling a bit sad or a bit moopy. This is what psychiatrists would diagnose as an illness.
Cathryn - That's right, absolutely. I mean, everyone has days or even weeks where they feel a bit under the weather, but we have very clear criteria for diagnosing depression that are well applied by GPs and beyond that by psychiatrists. So, we're really talking about severe episodes of depression that really stop people carrying out their normal life in the way that they would wish it.
Kat - In terms of where we are with understanding how genetics, how nature or nurture contribute to this? With the genetics part of it, where are we with trying to get a handle on the genes involved in depression?
Cathryn - So for depression, we've made less progress in identifying specific genes than many other diseases where research has moved very fast over the last few years. Now, we know from twin studies and from family studies that if someone has a relative with depression, then they themselves are at increased risk of depression. Genes have a major contribution to that. That's really interesting to know, but what we want to be able to do for the next step is identify the specific genes and the variants in those genes that could put people at increased risk of depression. There are several things that we can do with that. It may help us identify people that are at particular risk of depression before they suffer with an episode of depression. It gives us biological insights as to how depression works in the body, it may give us new drug targets and that would be hugely beneficial for the whole field.
Kat - In one of the talks we saw about the genetics of schizophrenia, they've done a lot of studies looking at lots and lots of people and you can kind of see this particular candidate genes popping out. That's not really the case with depression, is it? Why is that?
Cathryn - So, the schizophrenia studies have moved forward very fast and they now have over 100 different genes that they know play a role in schizophrenia. In depression, we haven't got that far. We have no confirmed genes for depression despite many decades of work for many international research groups. It seems that for depression, the whole disorder is much more heterogeneous. We know that people suffer from depression in different ways, different symptoms may be important to them, and that's clearly having an impact on how we can carry out genetic studies.
Kat - Is part of the problem that depression isn't just one thing, in the same way that cancer is now being divided, not just down into bowel cancer and breast cancer, but you have KRAS positive bowel cancer. Is the problem that we've just been putting everything that's depression into one bucket?
Cathryn - We suspect that that is true. We know that there are certain things in depression that make it more genetic, early onset depression, recurrent depression where people have subsequent episodes certainly have stronger family histories. It feels like depression, although it's very prevalent in the population, it's probably not a disorder with a single genetic pattern and it may be by subdividing different types of depression, looking at the different genes that contribute to subsets of that. And also, how that works with some of the environmental risk factors of things like stressful life events which we know increases risk of depression. I think we're going to need to model this more completely across all the different symptoms and the different contributions to risk before we can really make progress in identifying specific genetic variants for the disease.
Kat - And from your perspective, what one thing would you really like to see change in maybe the next couple of years in terms of our understanding of the biological causes of depression?
Cathryn - I'd like to see that we can identify some genes that really shed some light on the biological causes and what we can do about it. Just knowing the name of the genes doesn't get us very far. We need to take that downstream. We need to use that for developing more drugs for understanding the physiology of the disease. It's such a common disorder that we really should be making progress in this direction.
One of the things that we need to do this is we need large studies. We need people to get involved, to contribute DNA, to get involved in research when they're invited to do so. And it's really only with this combination between researchers like me, and patients and relatives suffering from the disease that we can hope to make progress.
Kat - That was Professor Cathryn lewis from Kings College London.

07:31 - Prof Mike Owen - Schizophrenia genes
Prof Mike Owen - Schizophrenia genes
with Professor Mike Owen, MRC Centre for Psychiatric Genetics and Genomics in Cardiff
Kat - We'll be returning to the hunt for depression-related genes later but now we're switching to a different mental health condition - schizophrenia. Affecting around one in a hundred people in the UK, schizophrenia can be extremely debilitating, as well as distressing for the family and friends of sufferers and a wider social issue too. Professor Mike Owen heads up the MRC Centre for Psychiatric Genetics and Genomics in Cardiff, and I asked him what we know so far about the genetic links to the disease.
Mike - So, we know that for example that if you have a first degree relative, a close relative with the disorder, say, a parent or a sibling, then your risk of getting the disorder is about 10 times increased. So, it's about one per cent generally in the population - a risk of getting schizophrenia. If you have a close relative, it's about 10 per cent. And we know that that could be shared genes, but it could be shared environments as well. It could be people living in the same sort of settings have similar experiences. So, you can do twin studies, adoption studies that show that that clustering in families, if you like, is largely due to shared genes. But there's a lot of evidence from those sorts of studies. More recently, now, there's many of us have been doing molecular genetics - looking at the DNA of people to try and identify the specific genes involved.
Kat - From your talk, one of the key problems was that schizophrenia isn't just one condition. It seems to be a wide range of conditions. How are some of the genetic studies starting to shed light on this complexity?
Mike - Well, the problem I guess is that what we call schizophrenia is a syndrome. It's a set of symptoms that occur together and it's a bit like trying to do genetics of shortness of breath, I guess, or of chest pain. Chest pain could be due to a heart attack or it could be due to stomach problems or muscular problems, and so forth. So yes, probably, it is heterogeneous. It contains various conditions. The other problem we have is that it's not nicely circumscribed. There are related disorders - bipolar disorder for example, which we hear a lot about in the press - kind of merges into it symptomatically. There's no kind of clear blue water between the two.
So, what the genetics is telling us is that genes for psychiatric disorders are not specific. The ones we've discovered so far are not generally speaking specific to one disorder. They're predisposed to multiple disorders, suggesting that these diagnostic categories that we use don't carve nature at the joints, if you like. There are reasonable sort of surface description of more complicated things going on lower down.
Kat - So, in terms of the genes that have come out on some of these studies, perhaps your top suspects, what do they tell us about some of the underlying biology of what's going on?
Mike - Well, the striking thing has been the degree of convergence onto sets of genes. Genes code for proteins, they contain the instructions for proteins. And so, what we can do is we can look at the proteins - the genes we found are coding for and see whether they cluster in particular functional groups. One of our most striking finding today is that there is a clustering of proteins that are involved in synapses.
Synapses are the sort of structures that allow nerve cells to communicate with each other. They're also the structures that basically confer the brain's ability to code information. So, the strength of synapses changes with experience and that codes information, if you like, and that plasticity is the basis for learning and memory, and helps the brain develop. So, we've identified genes that focus down on a small set of proteins that are involved in this synaptic plasticity.
Kat - And of course, what people with mental health problems, with schizophrenia, and their families will want to know is about, how will this knowledge help to treat the condition better. Where are we with coming up with more effective treatments. They don't seem to have been any breakthroughs for decades really.
Mike - No, there haven't really been any breakthroughs for 40 odd years. This work is very much aimed at trying to understand what is going wrong in the brains of people with severe psychiatric disorders and in the hope that we can identify new treatment targets. Now I have to say, this work is going to take a long time. The next step is to start to understand the function of some of these genes in the brain and we're starting to do that - well, in three ways.
Looking in people, we can look at people who carry specific genetic factors and we can study their brains with brain imaging methods. We can use animal models. We can manipulate the genes and model the risk genes in animals and study their brains and behaviour. A really exciting new technology is to use stem cells. We can take cells from people, skin cells and convert them into nerve cells and grow them in a dish and study their developments and the way they relate to other neurons, etc.
When you identify risk genes for disorders like this, the next phase is intensive kind of neuroscience and that's what's going to happen over the next five, 10, 15 years. I mean, we have to go this way. It's the only way I think to get effective treatments. It's very hard to predict how long anything in science will take. It could be a breakthrough next week, but I don't want to give false hope either, but there's nothing around the corner, but you know, this is solid progress. There's solid progress and I think we're entering a new phase of the game now.
Kat - That was Professor Mike Owen from the MRC Centre for Psychiatric Genetics and Genomics in Cardiff. And now it's time for a roundup of this month's genetics news.
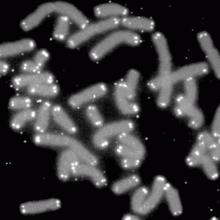
13:42 - Artificial chromosome created
Artificial chromosome created
In a major step forward in biological engineering, researchers have created the first 'designer' artificial yeast chromosome, publishing their landmark work in the journal Science. Brewer's yeast, Saccharomyces cerevisiae, has 16 chromosomes and shares 2,000 genes with humans. Previously researchers lead by Craig Venter have created synthetic chromosomes for bacteria, but yeast is a more complex organism with bigger chromosomes contained within a nucleus, similar to human cells.
Led by Dr Jef Boeke at New York University, the researchers made a synthetic version of yeast chromosome III, named SynIII, by designing and building nearly 275,000 base pairs of DNA - tens of thousands fewer than the number in the original chromosome. They cut the size down by removing repeated sections in the original DNA and some of the DNA that doesn't encode instructions for making proteins. Despite having more than 50,000 changes to its DNA code, the designer yeast were still able to grow and multiply as usual. It also gained new functions, such as the ability to scramble the new chromosome on cue to make future genetic manipulations easier.
The new designer chromosome is the first step in an international project aiming to build yeast's entire genome over the next few years. Scientists are also developing synthetic genes that would transform yeast into biological factories, for example producing antibiotics and turning agricultural waste into biofuel.
The researchers also think that it might one day be feasible to synthesise entire plant and animal chromosomes, or even whole genomes. That may be a long way off, but certainly mini chromosomes containing tens or even hundreds of genes are definitely within the foreseeable future, according to the team.
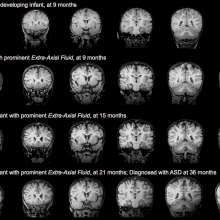
15:10 - Autism begins before birth?
Autism begins before birth?
Writing in the New England Journal of Medicine, scientists from the University of California say they have new evidence that autism begins in the womb. The conclusions come from studying brain tissue from 22 children with and without autism that had died in accidents or from unrelated diseases, aged between 2 and 15 years. The scientists discovered key differences in the outer layer of the brain, called the cortex, in ten out of 11 children with autism, compared to one in 11 children without the condition.
The changes were dotted about in brain regions involved in social and emotional communication, and language, and are likely to have arisen during the second or third trimester of pregnancy. Importantly, the researchers think that their findings might explain why some toddlers with autism show signs of improvement if treated early enough, as younger brains are more likely to rewire themselves. So the race is on to find out whether these changes hold up in further studies, and how the information might be used to inform earlier identification of autistic children so appropriate interventions can be made as early as possible.

16:12 - Gene for baldness found
Gene for baldness found
From snake oil creams to hair transplants, men around the world are keen for a cure for baldness. But while there's still no cure for male pattern baldness, researchers have made a step forward in understanding the gene fault lying behind a rare balding condition called "atrichia with papular lesions".
Writing in The FASEB Journal, US researchers have shown that a gene called Hairless plays a crucial role in controlling when other genes are switched on and off. It does this by removing tiny molecular tags, called methyl groups, from the proteins that package up DNA, known as histones. The scientists hope their discovery will open up new avenues for developing treatments for rare forms of hair loss as well as skin diseases including skin cancer.

17:08 - Prof Jonathan Flint - Melancholic genes
Prof Jonathan Flint - Melancholic genes
with professor Jonathan Flint, Oxford University
Kat - Continuing our reports from the Genetics Society Spring meeting, focusing on the genetics underlying psychiatric disorders, it's time to hear from the winner of the annual Genetics Society's Medal, Professor Jonathan Flint from the University of Oxford. At the meeting he presented fascinating early results from his team's hunt for genes involved in major depression, and told me more about the study.
Jonathan - So we thought, along with other people, that the most critical thing to do is to try and reduce heterogeneity because there are many different ways that you could become depressed - many different causes, both environmental and genetic. Unless you try and reduce heterogeneity then your analysis is either going have to be so large as probably to be unwieldy and we felt also that it's very difficult to understand what the interplay and interacting factors might be. So, the study we designed was to hone in on a group where we thought that the causes would be as homogeneous as possible.
So for example, we know that the genetic effects are different in men and women and we know that women have higher rates of depression than men. So, it seems sensible just to pick one sex and we chose women. And we know that recurrent depression is more likely to be heritable than non-recurrent depression, so we choose heritable depression. And we know the causes are slightly different if your onset is very, very early or later in life. So, we restrict the age categories at which the depression occurs.
Kat - And as well with your study, you didn't carry this out in the UK. Tell me about where you did it.
Jonathan - No, we didn't carry it out in UK. So, because we have to go through all of these restrictions, although the disorder is common - so if you just took one episode of depression as a measure of the occurrence in a lifetime - the prevalence rate is something between 10 to 15 per cent. So, it's pretty common. But because we've restricted it to recurrent and only one sex, and a particular age group, and they have to have certain other features, that made it rarer. And we also require interviewers to spend a long time with their patients. So, the number of people who were prepared to give up that time is of course, even less.
We wanted it to be genetically homogeneous because we're very interested in the genetic causes. All of those features rather restricted where we could do the study. In the end, we decided to try China because they're obviously an enormous population and it has a very good healthcare system, and the doctors are very well-trained. We can relatively restrict genetic homogeneity by only going for Han Chinese, which is what we did.
Kat - Tell me about the size of the study. How many women were you looking at?
Jonathan - We decided to choose 6,000 cases and 6,000 controls as our target. We made that decision about 2006, 2007, so about seven years ago now. On the small side in fact for what we now know. And ideally, we'd like probably 10 times that number. You can argue about this but we thought by restricting it, we'd have a chance of finding something. By May of 2012, we had indeed reached our sample of 6,000 cases and 6,000 controls.
Kat - So, as well as interviewing these women and getting the doctors to talk to them and find out, you were also looking at their genes as well.
Jonathan - We sequenced everybody. We have their genome sequences in a large database now, yes.
Kat - And so, can you tell me about some of the things that you've started to find? What seems to be coming out from this data?
Jonathan - Well, so far, but this is a little preliminary, we've discovered - we think - one locus in the genome that's more likely to make people depressed.
Kat - So, that's one kind of genetic area, one gene.
Jonathan - Yes and it's clearly not the only one. This is a very small effect and everything tells us, including our own data, that it's highly polygenic. But there's only one which reaches accepted statistical significance and we've yet to replicate that. But it's an intriguing observation because it lies over a gene called Sirtuin-1. That's intriguing because that gene has been implicated in the a whole series of phenotypes, many of which are indeed related to the phenotypes of depression. So, for example, depressed people typically lose their appetite. They don't sleep very well. They become a little sluggish. And knockouts with that gene also manifest many of those phenotypes. And perhaps most importantly, the Sirtuin-1 locus has been regarded as at least a moderator of metabolic phenotypes. One way of conceptualising depression is that there's a certain form of it called melancholia, in which by these so-called vegetative symptoms predominate.
Kat - So, these sound like quite intriguing links. What next for this study? How do you go about trying to work out if this is actually true?
Jonathan - So, the thing we'd like to do most of all is replicate and we've been scouring the world's genetics groups to see if they've got a similar sample to ours. The problem has been, because we've been so restricted in what we collect, no one has got a similar sample. So, we have a little problem there. The alternative is for us to collect another sample, so we're trying to do that as well. But the other thing that we can do is we can use our sequence data in rather interesting ways.
So, when people normally carry out these studies, they interrogate the genome at maybe one million positions and these are typically those which vary with frequencies of greater than five per cent. But we've got sequence data and we can almost interrogate everything. Not quite because of the way we've done it. So, that allows us to look at the genes we think might be involved from the genome-wide association study and look for other signals. So, one of the signals that we look for is whether there might be small deletions, mutations in the genes that we think are involved and now, and are they enriched in cases or controls. There is a little hint that indeed might be so. So, that provides an independent signal, because it's not incorporated in the genome-wide association, that what we found is real.
Kat - It seems to be that with some diseases such schizophrenia, there's a lot of genetic data coming out with autism as well. Depression seems a bit of an impenetrable box. Where do you think this is going to be heading and do you hope to find a hatful of genes, or how do you see the field progressing?
Jonathan - We could, as I think most people would agree, go ahead by collecting a larger sample. There's little doubt that if you collect a big enough sample, you'll find more things. There's a quite interesting linear correlation with the number of loci you find in the sample size. So in a simple way, if you double your sample size, you'll double the number of loci that you find. So, if we find with 6,000, one locus, we go up to 60,000, we'll find 10. So, that's one simple approach one could take. One could refine the phenotype further and the information we're collecting already tells us that's an important way to go.
Kat - So, just picking people with very, very specific symptoms.
Jonathan - Yes. So maybe, for example, what the locus we found is specific for highly recurrent depression and for a subtype of depression called melancholia which is, if you like, some sort of measure the severity. And it might be possible to subdivide it further. Once we know what type of depression shows these features, shows the genetic linkage, the genetic association, then we could go ahead and specifically look for those. That's one thing that we could do.
Kat - And more broadly, with the kind of studies that we've been hearing about in the meeting, looking more in-depth at the genetics, the underlying causes of some of these mental disorders, where do you think perhaps in five or 10 years we might be with understanding some of these really complex and very challenging, and in many cases, distressing illnesses?
Jonathan - Well, I think we have to take the genetic findings for what they are - namely, that it's unbiased approach to the underlying biology. And while that's good in a sense that, for most psychiatric conditions, probably for all, we don't really understand what the causes are. So, any clue is good. On the other hand, it means that there's an awful lot of work in turning a genetic signal into what we all want which is mechanism and possibly later some way of treating that condition.
Kat - That was Professor Jonathan Flint from the University of Oxford.

Gene therapy for Huntington's Disease?
Kat - And here's Harriet Johnson with this month's listener question. Harriet - Listener Cari says, "This year, I learned I'm gene positive for Huntington's disease. I'm relatively young, so I'd like to know what is the latest research and realistic hope in terms of a genetic therapy as a cure for me." To answer, here's Nobel Prize winner, Craig Mello from the Howard Hughes Medical Institute at the University of Massachusetts.
Craig - That is a very difficult question to answer. Right now, these therapies are based on RNA interference which uses the body's own small RNA silencing system. Every cell in our bodies has basically a search engine system, much like we use for browsing the internet. Several years ago, scientists learned how to enter search queries into that search engine and when this search engine finds a target, it can use the sequence information to very precisely identify a piece of the genetic information and turn it off. And now, therapies are now beginning to move into the clinic that are based on entering search queries into the cell's own searching machinery and then directing it to a desired target - in this case, the Huntington's gene. It's not a cure. It's a therapy and the way that this is being delivered would be using a viral vector. In this case, we put a virus to work for us. It delivers into the brain cell directly a small gene that expresses the RNA that enters into this search query machinery I told you about. So, we sort of hijack the cell's own machinery to try to turn off this disease gene. Now, that of course is going to need to be tested further in animals. So there are safety concerns that need to be tested - that's going to take a few more years. Then it will enter clinical trials - that'll take a couple of years, maybe more, then gradually, into bigger clinical trials. So, we're talking about maybe a 10-year horizon before you see a lot of people potentially getting this therapy. There are additional new things coming such as this new technology called CRISPR. One of the benefits of CRISPR would be that it could permanently destroy and inactivate the diseased allele. However, there are a lot of safety concerns with CRISPR in getting it into the cells. Delivery is even more challenging. It's a totally different mechanism that's not endogenous even to human cells. It's something that bacteria have and we're going to have to put the machinery for CRISPR into the human in order to do this. So, this is years away.
Kat - Thanks to Professor Craig Mello for that answer. And if you've got any questions about genes, DNA and genetics, just email them to me at genetics@thenakedscientists.com.

28:13 - Gene of the month - Toddler
Gene of the month - Toddler
with Kat Arney
Kat - And now, it's time for our Gene of the Month - and this time it's Toddler. Officially known as Apela, Toddler is a new addition to the genetic canon, having only been discovered earlier this year. It was identified in zebrafish, and plays a vital role in very early development, promoting the movement of cells as a fish embryo transforms from a tiny football of cells into a more folded structure with a head and tail. Without Toddler, cells move sluggishly, resulting in embryos with small hearts or no heart at all.
Researchers had thought they were aware of pretty much all the genes involved in early development, so Toddler's appearance was a surprise. It had gone under the radar for so long as the protein encoded by the gene is very small, and scientists simply hadn't noticed it. It's possible that there may be a wealth of other tiny genes running around out there, waiting to be discovered.
- Previous Why do we laugh when tickled?
- Next The first in-vitro vagina?










Comments
Add a comment